 3D rendering of COVID-19 virus particles. (Photo by iStock)
3D rendering of COVID-19 virus particles. (Photo by iStock)Researchers devise a new way to detect and quantify SARS-CoV-2 in pooled samples
Testing wastewater for SARS-CoV-2 is one way public health departments are monitoring the spread of COVID-19 within their communities, but dilution in these large, pooled samples can prevent accurate readings of viral levels. Motivated by this challenge, UC Berkeley researchers have developed a new, more sensitive method for detecting and quantifying this virus in large sample volumes.
In a study published in ACS Analytical Chemistry, Lydia Sohn, the Almy C. Maynard and Agnes Offield Maynard Chair in Mechanical Engineering, and her research team demonstrated how their innovative test method, dubbed DIVER, can provide clearer insight into a population’s active COVID infections than current approaches.
“DIVER offers two key advantages over RT-qPCR [reverse transcription quantitative polymerase chain reaction] testing, which is considered the gold standard for SARS-CoV-2 detection,” said Sohn. “It can detect intact virus versus RNA fragments, and, more importantly, DIVER is effective in almost any sample volume size, making it perfect for pooled samples.”
These advantages allow DIVER to more accurately gauge the prevalence of active COVID-19 infections. Intact virus has been shown to be a better indicator of current viral infection than RNA fragments, which are shed in bodily fluids by people who have had COVID but who may no longer be infectious. And because DIVER is sensitive enough to detect low concentrations of intact virus in even large sample volumes, communities can benefit from the efficiency of testing pooled samples without compromising on sensitivity.
DIVER, which stands for Detection of Intact Virus by Exogenous-nucleotide Reaction, is a five-step testing process. Researchers begin by attaching oligos — which are short sequences of DNA strands — to the outer layer of the virus. Next, they isolate the virus using magnetic beads coated with ACE2, a special protein found in the human body that likes to bind to the SARS-CoV-2 spike protein. In this way, ACE2 acts like Velcro for the virus particles.
Once the virus is attached to the ACE2-coated beads and isolated, researchers use a technique called quantitative polymerase chain reaction to determine the number of intact virus particles and the level of active infection within the sample group.
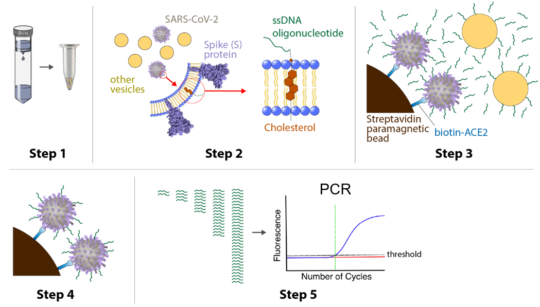
DIVER uses a five-step process to identify, isolate and quantify SARS-CoV-2 virus particles in large sample volumes. Researchers take their sample and attach oligos — short sequences of DNA strands — to the outer layer of the virus (steps 1 and 2). Then they isolate the virus using magnetic beads coated with ACE2, a special protein found in the human body that likes to bind to the SARS-CoV-2 spike protein (step 3). Once the virus is attached to the ACE2-coated beads and isolated (step 4), researchers use a technique called quantitative polymerase chain reaction to quantify the intact virus particles (step 5). (Image by Thomas Carey)
According to Sohn, DIVER’s use of DNA as a tracking tool makes it easier to find the virus, a major challenge when testing large, pooled samples like wastewater. “Since each virus particle is tagged with many copies of the same DNA oligos, our sensitivity is greatly increased,” said Sohn. “This is in comparison to the standard, where detection is based on a single copy of the RNA found in the virus.”
DIVER could be used to detect and quantify a wide range of viruses, not just known strains of COVID-19. Users can swap out ACE2 for a Velcro-like capture agent specific to a different virus, as long as the target virus is enclosed by a lipid membrane.
“This makes DIVER adaptable to any emerging SARS-CoV-2 variants or any enveloped virus,” said Sohn.
